Note: I am experimenting with exporting Jupyter notebooks into a WordPress ready format. This notebook refers specifically to to the Nature Conservancy Kaggle, for classifying fish species, based only on photographs.
Loading
better living through computation.
Note: I am experimenting with exporting Jupyter notebooks into a WordPress ready format. This notebook refers specifically to to the Nature Conservancy Kaggle, for classifying fish species, based only on photographs.
While studying Operating Systems, I decided to experiment with 4 of the most common Page Replacement algorithms, via simulation, using Python. Input data for my simulations were two memory traces named “swim.trace” and “gcc.trace” (provided by my CS department). All algorithms are run within a page table implemented for a 32-bit address space; all pages in this page table are 4kb in size.
I chose to implement these algorithms and the page table in Python. The final source code can be found here, on Github. The full data set that I collected can be found at the end of this document. Illustrative graphs are interspersed throughout, wherever they are necessary for explaining and documenting decisions.
In this essay itself, I will first outline the algorithms implemented, noting design decisions I made during my implementations. I will then compare and contrast the results of each algorithm when run on the two provided memory traces. Finally, I will conclude with my decision about which algorithm would be best to run in a real Operating System.
Please note, to run the algorithms themselves, you must run:
The algorithms we have been asked to implement are:
The main entry point for the program is the file named vmsim.py. This is the file which must be invoked from the command line to run the algorithms. Because this is a python file, it must be called with “python vmsim.py … etc.”, instead of “./vmsim”, as a C program would. Please make sure the selected trace file is in the same directory as vmsim.py.
All algorithms run within a page table, the implementation of which can be found in the file named pageTable.py.
Upon program start, the trace file is parsed by the class in the file parseInput.py, and each memory lookup is stored in a list of tuples, in this format: [(MEM_ADDRESS_00, R/W), [(MEM_ADDRESS_01, R/W), … [(MEM_ADDRESS_N, R/W)]. This list is then passed to whichever algorithm is invoked, so it can run the chosen algorithm on the items in the list, element by element.
The OPT algorithm can be found in the file opt.py. My implementation of OPT works by first preprocessing all of the memory addresses in our Trace, and creating a HashTable where the key is our VPN, and the value is a python list containing each of the address numbers at which those VPNs are loaded. Each time a VPN is loaded, the element at index 0 in that list is discarded. This way, we only have to iterate through the full trace once, and from there on out we just need to hash into a list and take the next element, whenever we want to know how far into the future that VPN is next used.
The Clock Algorithm is implemented in the files clock.py and circularQueue.py. My implementation uses the second chance algorithm with a Circular Queue. Of importance: whenever we need to make an eviction, but fail to find ANY pages that are clean, we then run a ‘swap daemon’ which writes out ALL dirty pages to disk at that time. This helps me get fewer page faults, at the expense of more disk writes. That’s a calculated decision for this particular algorithm, since the project description says we should use page faults as our judgment criterion for each algorithm’s effectiveness.
The LRU Algorithm can be found in the file lru.py. For LRU, each time a page is Read, I mark the memory address number at which this happens in the frame itself. Then, in the future—whenever I need to make an eviction—I have easy access to see which frame was used the longest time in the past, and no difficult calculations are needed. This was the simplest algorithm to implement.
The Aging Algorithm is likely the most complex, and its source code can be found in the file named aging.py. Aging works by keeping an 8 bit counter and marking whether each page in the page table was used during the last ‘tick’ a time period of evaluation which must be passed in by the user as a ‘refresh rate’, whenever the Aging Algorithm is selected. All refresh rates are in milliseconds on my system, but this relies on the implementation of Python’s “time” module, so it’s possible that this could vary on other systems. For aging, I suggest a refresh rate of 0.01 milliseconds, passed in on the command line as “-r 0.01”, in the 2nd to last position in the arg list. This minimizes the values in my testing, and going lower does not positively affect anything. In the next section, I will show my rationale for selecting 0.01 as my refresh rate.
In order to find a refresh rate that would work well, I decided to start at 1ms and move 5 orders of magnitude in each direction, from 0.00001ms to 100000ms.
To ensure that the results were not biased toward being optimized for a single trace, I tried both to confirm that the refresh rate would work will for all inputs.
The graphs below show the Page Faults and Disk Writes I found during each test.
For all tests, I chose a frame size of 8, since this small frame size is most sensitive to the algorithm used. At higher frame sizes, all of the algorithms tend to perform better, across the board. So I wanted to focus on testing at the smallest possible size, preparing for a ‘worst case’ scenario.

X-axis: Refresh rate in milliseconds
Y-axis: Total Page faults
GCC. TRACE reaches its minimum for page faults at 0.0001ms and SWIM.TRACE reaches its own minimum at 0.01ms. The lines cross at around 0.01ms.

X-axis: Page Faults
Y-Axis: Refresh Rates

X-axis: Page Faults
Y-Axis: Refresh Rates
Additionally, 0.01ms seems to achieve the best balance, in my opinion, if we need to select a single time for BOTH algorithms.

X-axis: Refresh rate in milliseconds
Y-axis: Total Disk Writes
The number of disk writes also bottoms out at 0.01ms from SWIM.TRACE. It is relatively constant for GCC.TRACE across all of the different timing options.
Because of this, I suggest 0.01ms as the ideal refresh rate. This is because it is optimal for SWIM.TRACE. For GCC.TRACE, it is not the absolute best option, but it is still acceptable, and so I think this selection will achieve a good balance.
With the algorithms all implemented, my next step was to collect data for each algorithm at all frame sizes, 8, 16, 32, and 64. OPT always performed best, and thus it was used as our baseline.
In the graphs below, I show how each algorithm performed, both in terms of total page faults and total disk writes.

X-axis: Frame Size
Y-axis: Page Faults
Data for all algorithms processing swim.trace

X-axis: Frame Size
Y-axis: Disk Writes
Data for all algorithms processing swim.trace

X-axis: Frame Size
Y-axis: Page Faults
Data for all algorithms processing gcc.trace

X-axis: Frame Size
Y-axis: Disk Writes
Data for all algorithms processing gcc.trace
—
Given this data, I was next tasked with choosing which algorithm is most appropriate for an actual operating system.
In order to determine which algorithm would be best, I decided to use an algorithm. I’ll call it the ‘Decision Matrix’, and here are the steps.
DECISION MATRIX:
| ALGORITHM | SWIM – Page Faults | SWIM – Disk Writes | GCC – Page Faults | GCC – Disk Writes | TOTAL (Lowest is Best) |
| OPT | 1 | 1 | 1 | 1 | 4 |
| CLOCK | 2 | 4 | 2 | 4 | 12 |
| AGING | 4 | 2 | 4 | 2 | 12 |
| LRU | 3 | 3 | 3 | 3 | 12 |
Ranking: 1=Best;
4=Worst
NOTE:
Therefore, I would select LRU for my own operating system.
Figure 1.1 – Full Data Set
| ALGORITHM | NUMBER OF FRAMES | TOTAL MEMORY ACCESSES | TOTAL PAGE FAULTS | TOTAL WRITES TO DISK | TRACE | REFRESH RATE |
| OPT | 8 | 1000000 | 236350 | 51162 | swim.trace | N/A |
| OPT | 16 | 1000000 | 127252 | 27503 | swim.trace | N/A |
| OPT | 32 | 1000000 | 52176 | 11706 | swim.trace | N/A |
| OPT | 64 | 1000000 | 24344 | 6316 | swim.trace | N/A |
| OPT | 8 | 1000000 | 169669 | 29609 | gcc.trace | N/A |
| OPT | 16 | 1000000 | 118226 | 20257 | gcc.trace | N/A |
| OPT | 32 | 1000000 | 83827 | 14159 | gcc.trace | N/A |
| OPT | 64 | 1000000 | 58468 | 9916 | gcc.trace | N/A |
| CLOCK | 8 | 1000000 | 265691 | 55664 | swim.trace | N/A |
| CLOCK | 16 | 1000000 | 136154 | 52104 | swim.trace | N/A |
| CLOCK | 32 | 1000000 | 73924 | 45872 | swim.trace | N/A |
| CLOCK | 64 | 1000000 | 56974 | 43965 | swim.trace | N/A |
| CLOCK | 8 | 1000000 | 178111 | 38992 | gcc.trace | N/A |
| CLOCK | 16 | 1000000 | 122579 | 26633 | gcc.trace | N/A |
| CLOCK | 32 | 1000000 | 88457 | 20193 | gcc.trace | N/A |
| CLOCK | 64 | 1000000 | 61832 | 15840 | gcc.trace | N/A |
| AGING | 8 | 1000000 | 257952 | 52664 | swim.trace | 0.01ms |
| AGING | 16 | 1000000 | 143989 | 41902 | swim.trace | 0.01ms |
| AGING | 32 | 1000000 | 91852 | 29993 | swim.trace | 0.01ms |
| AGING | 64 | 1000000 | 82288 | 27601 | swim.trace | 0.01ms |
| AGING | 8 | 1000000 | 244951 | 31227 | gcc.trace | 0.01ms |
| AGING | 16 | 1000000 | 187385 | 22721 | gcc.trace | 0.01ms |
| AGING | 32 | 1000000 | 161117 | 19519 | gcc.trace | 0.01ms |
| AGING | 64 | 1000000 | 149414 | 16800 | gcc.trace | 0.01ms |
| LRU | 8 | 1000000 | 274323 | 55138 | swim.trace | N/A |
| LRU | 16 | 1000000 | 143477 | 47598 | swim.trace | N/A |
| LRU | 32 | 1000000 | 75235 | 43950 | swim.trace | N/A |
| LRU | 64 | 1000000 | 57180 | 43026 | swim.trace | N/A |
| LRU | 8 | 1000000 | 181950 | 37239 | gcc.trace | N/A |
| LRU | 16 | 1000000 | 124267 | 23639 | gcc.trace | N/A |
| LRU | 32 | 1000000 | 88992 | 17107 | gcc.trace | N/A |
| LRU | 64 | 1000000 | 63443 | 13702 | gcc.trace | N/A |
Figure 1.2 – GCC.TRACE – Refresh Rate Testing – 8 Frames
| ALGORITHM | NUMBER OF FRAMES | TOTAL MEMORY ACCESSES | TOTAL PAGE FAULTS | TOTAL WRITES TO DISK | TRACE | REFRESH RATE |
| AGING | 8 | 1000000 | 192916 | 33295 | gcc.trace | 0.00001ms |
| AGING | 8 | 1000000 | 192238 | 33176 | gcc.trace | 0.0001ms |
| AGING | 8 | 1000000 | 197848 | 31375 | gcc.trace | 0.001ms |
| AGING | 8 | 1000000 | 244951 | 31227 | gcc.trace | 0.01ms |
| AGING | 8 | 1000000 | 339636 | 140763 | gcc.trace | 0.1ms |
Figure 1.3 –SWIM.TRACE – Refresh Rate Testing – 8 Frames
| ALGORITHM | NUMBER OF FRAMES | TOTAL MEMORY ACCESSES | TOTAL PAGE FAULTS | TOTAL WRITES TO DISK | TRACE | REFRESH RATE |
| AGING | 8 | 1000000 | 275329 | 53883 | swim.trace | 0.00001ms |
| AGING | 8 | 1000000 | 274915 | 53882 | swim.trace | 0.0001ms |
| AGING | 8 | 1000000 | 268775 | 53540 | swim.trace | 0.001ms |
| AGING | 8 | 1000000 | 257952 | 52664 | swim.trace | 0.01ms |
| AGING | 8 | 1000000 | 278471 | 56527 | swim.trace | 0.1ms |
Example code to start experimenting with the Harris Corner Detection Algorithm on your own, in Matlab. And some of the results I obtained in my own testing.
Among the classic algorithms in Computer Vision is Harris Corner Detection. (For the original paper, from 1988, see here.) The problem it solves: Given any image file, can you find which areas correspond to a corner with a high degree of certainty?
The answer is yes. And the algorithm is elegant. Essentially the steps are:
Recently, I was trying to implement my own version of the Harris Detector in Matlab, and ended up banging my head against the wall for a few hours while figuring out some of the subtler details the hard way.
Here’s the code I came up with, and some examples of the outputs. Grab the code from both gists below, and you can start experimenting on your own.
| function [ x, y, scores, Ix, Iy ] = harris_corners( image ) | |
| %HARRIS_CORNERS Extracts points with a high degree of 'cornerness' from | |
| %RGB image matrix of type uint8 | |
| % Input – image = NxMx3 RGB image matrix | |
| % Output – x = nx1 vector denoting the x location of each of n | |
| % detected keypoints | |
| % y = nx1 vector denoting the y location of each of n | |
| % detected keypoints | |
| % scores = an nx1 vector that contains the value (R) to which a | |
| % a threshold was applied, for each keypoint | |
| % Ix = A matrix with the same number of rows and columns as the | |
| % input image, storing the gradients in the x-direction at each | |
| % pixel | |
| % Iy = A matrix with the same nuimber of rwos and columns as the | |
| % input image, storing the gradients in the y-direction at each | |
| % pixel | |
| % compute the gradients, re-use code from HW2P, use window size of 5px | |
| % convert image to grayscale first | |
| G = rgb2gray(image); | |
| % convert to double | |
| G2 = im2double(G); | |
| % create X and Y Sobel filters | |
| horizontal_filter = [1 0 -1; 2 0 -2; 1 0 -1]; | |
| vertical_filter = [1 2 1; 0 0 0 ; -1 -2 -1]; | |
| % using imfilter to get our gradient in each direction | |
| filtered_x = imfilter(G2, horizontal_filter); | |
| filtered_y = imfilter(G2, vertical_filter); | |
| % store the values in our output variables, for clarity | |
| Ix = filtered_x; | |
| Iy = filtered_y; | |
| % Compute the values we need for the matrix… | |
| % Using a gaussian blur, because I get more positive values after applying | |
| % it, my values all skew negative for some reason… | |
| f = fspecial('gaussian'); | |
| Ix2 = imfilter(Ix.^2, f); | |
| Iy2 = imfilter(Iy.^2, f); | |
| Ixy = imfilter(Ix.*Iy, f); | |
| % set empirical constant between 0.04-0.06 | |
| k = 0.04; | |
| num_rows = size(image,1); | |
| num_cols = size(image,2); | |
| % create a matrix to hold the Harris values | |
| H = zeros(num_rows, num_cols); | |
| % % get our matrix M for each pixel | |
| for y = 6:size(image,1)-6 % avoid edges | |
| for x = 6:size(image,2)-6 % avoid edges | |
| % calculate means (because mean is sum/num pixels) | |
| % generally, this algorithm calls for just finding a sum, | |
| % but using the mean makes visualization easier, in my code, | |
| % and it doesn't change which points are computed to be corners. | |
| % Ix2 mean | |
| Ix2_matrix = Ix2(y-2:y+2,x-2:x+2); | |
| Ix2_mean = sum(Ix2_matrix(:)); | |
| % Iy2 mean | |
| Iy2_matrix = Iy2(y-2:y+2,x-2:x+2); | |
| Iy2_mean = sum(Iy2_matrix(:)); | |
| % Ixy mean | |
| Ixy_matrix = Ixy(y-2:y+2,x-2:x+2); | |
| Ixy_mean = sum(Ixy_matrix(:)); | |
| % compute R, using te matrix we just created | |
| Matrix = [Ix2_mean, Ixy_mean; | |
| Ixy_mean, Iy2_mean]; | |
| R1 = det(Matrix) – (k * trace(Matrix)^2); | |
| % store the R values in our Harris Matrix | |
| H(y,x) = R1; | |
| end | |
| end | |
| % set threshold of 'cornerness' to 5 times average R score | |
| avg_r = mean(mean(H)); | |
| threshold = abs(5 * avg_r); | |
| [row, col] = find(H > threshold); | |
| scores = []; | |
| %get all the values | |
| for index = 1:size(row,1) | |
| %see what the values are | |
| r = row(index); | |
| c = col(index); | |
| scores = cat(2, scores,H(r,c)); | |
| end | |
| y = row; | |
| x = col; | |
| end |
| image = imread('your_image.png'); | |
| image = imresize(image, 0.75); # or however much you want to resize if your image is large | |
| [ x, y, scores, Ix, Iy ] = harris_corners( image ); | |
| figure; imshow(image) | |
| hold on | |
| for i = 1:size(scores,2) | |
| plot(x(i), y(i), 'ro', 'MarkerSize', scores(i) * 2); # you may need to play with this multiplier or divisor based on your image | |
| # I've used –> (/1000) to (* 10) | |
| end | |
| saveas(gcf,'your_image_with_corners.png'); | |
| hold off |
Here’s what you can expect to see. I’m coloring the corners RED, using circles that grow larger the more likely the area is to hold a corner. So, larger circles –> more obvious corners. (Also note that I’m downsizing my output images slightly, just to make computation faster. )
That’s it. With this code, you can start experimenting with Harris Detectors in about 30 seconds. And hopefully the way I’ve written it is transparent enough to make it obvious what’s going on behind the scenes. Most of the implementations out there are somewhat opaque and hard to understand at a conceptual level. But I’m hoping that my contribution will make this all a little more straightforward to those just gettings started. Enjoy. -t
I’m currently working on a computer vision project to implement seam carving, which is explained quickly in this video by the technique’s inventors. The goal of seam carving is to allow for content-aware image resizing. Part of this algorithm is a numerical method to identify the ‘important’ content of an image (at least w/r/t re-sizing). I wanted to see what this algorithm would ID as ‘important’ in some great artworks from history. So I ran the algorithm on some of my favorite paintings (low-res versions from the internet) and am sharing the outputs here.
The short story is that seam carving allows us to re-size images without distorting the important details. Seam carving is the process of finding connected paths of pixels with “low energy” running through the image–call these paths seams–and then removing those seams instead of a full column or row which might intersect with something important in the image, like a person’s face, the edge of a large object, etc… The easiest way to get an overview is to watch the video above, but hopefully that’s a succinct enough explanation for anyone impatient like me.
The first step in implementing the seam carving algorithm is to compute what’s called the energy function of the image’s pixel gradients. We can do this by plotting the image’s grayscale values (0-255) in a 2D matrix, applying a filter to identify edge gradients in both the x-direction and y-direction, and then computing the magnitude of each pixel’s edge gradient, weighing the x-gradient and y-gradient equally: (i.e. — sqrt(x^2 + y^2) ). If that doesn’t make a ton of sense yet, don’t worry. The video is the easiest way to understand the concept, and reading the paper is the easiest way to understand the process. This is just the super succinct version. (I also probably explained it badly.)
| function [ energy_matrix ] = energy_image( image_matrix_input ) | |
| %ENERGY_IMAGE Computes the energy at each pixel in a matrix nxmx3 matrix | |
| % Outputs a 2D-matrix containing energy equation outputs, of datatype DBL | |
| % convert image to grayscale first | |
| G = rgb2gray(image_matrix_input); | |
| % convert to double | |
| G2 = im2double(G); | |
| % create X and Y filters | |
| horizontal_filter = [1 0 -1; 2 0 -2; 1 0 -1]; | |
| vertical_filter = [1 2 1; 0 0 0 ; -1 -2 -1]; | |
| % using imfilter to get our gradient in each direction | |
| filtered_x = imfilter(G2, horizontal_filter); | |
| filtered_y = imfilter(G2, vertical_filter); | |
| energy_matrix = zeros(size(G2,1), size(G2,2)); | |
| % compute the energy at each pixel using the magnitude of the x and y | |
| % gradients: sqrt((dI/dX)^2+(dI/dY)^2)) | |
| for y = 1:size(G2, 1) | |
| for x = 1:size(G2,2) | |
| % calculate energy function | |
| y_magnitude = filtered_y(y,x) ^ 2; | |
| x_magnitude = filtered_x(y,x) ^ 2; | |
| energy_output = sqrt( y_magnitude + x_magnitude ); | |
| % fill energy matrix with our calculation | |
| energy_matrix(y,x) = energy_output; | |
| end | |
| end | |
| end |
Okay, so now we can compute the energy function of any image.
And the supposed output of this energy function is a mapping where the the important details of the image–the ones we don’t want to carve out when we re-size–are represented in higher intensities of white. The less important details of the image are represented in darker shades of gray and black.
So my question is what happens when we run historical artworks through this algorithm? Does it tell us anything new and novel about artist, his techniques, etc? I’ve always heard that the really well educated and expert art historians can confirm or deny the veracity of an artwork by looking at brush strokes, and the like. I’ve also read stories about taking x-rays of famous artworks, and revealing hidden paintings beneath.
For the most part I’ll let you decide, but in any case, I found some of the outputs to be really beautiful and interesting in their own right. Enough talking for now, here’s the imagery.
You’ll have to click through to see the images in full-size (bottom right button in popup after clicking on the thumbnail), and some of the more interesting details need magnification, which requires downloading the images, just as a heads up.
There’s a lot more that can be done with this, of course, and this post has no real analysis, conclusions, or anything of the like. But I wanted to share what I was playing around with, in case anyone else finds it interesting, like I do. -t
I needed to generate Standard Normal Random Variates for a simulation I’m programming, and I wanted to see which method would be best to use. There are a few options, but generally you need to experiment to know for sure which will work best in your situation.
I decided to do this in Python, and to share my code + experimentation process and recommendations/reasoning.
Hoping this can save someone else some time later. If you need to do the same for your own simulation, or any other reason, hopefully this solves some of your problems, helps you understand what’s going on under the hood, and just makes life easier.
Grab the code here: https://github.com/adpoe/STD-Normal-Rand-Variates/tree/master/code
Experimental Setup
I used Python to algorithmically generate random variates that follow the Standard Normal distribution according to three different methods. For all methods, 10,000 valid random variables were generated in each algorithm’s run, in order to maintain consistency for later effectiveness comparisons. The methods tested were:
In the following paragraphs, I will briefly outline the implementation decisions made to generate Standard Normal random variates according to each method. I will then analyze the results, compare them, and issue my own recommendation on which method to use going forward, informed by the data gathered in this experiment.
Inverse Transform
The Inverse Transform Method works by finding the inverse of the CDF for a given probability distribution (F-1(X)), then feeding random numbers generated from U[0,1) into that inverse function. This will yield randomly generated variables within the range of our desired probability distribution.
However, this method is problematic for the Standard Normal Distribution, because there is no closed form for its CDF, and hence we cannot calculate its exact inverse. Because of this, I chose to use Bowling’s closed-form approximation of the Standard Normal CDF, which was developed in 2009: Pr(Z <= z) = 1 / [1 + e^(-1.702z)].
Despite being only an approximation, Bowling’s closed form CDF function is mathematically close enough to generate reasonable random variates. Beyond that, this function is simple. The hardest part was calculating the inverse, which was actually done with help from Wolfram Alpha. Once an inverse was obtained, implementation was straightforward and can be seen in the code attached, within the method @inverse_transform().
Accept/Reject
The Accept/Reject method for random variate is more complex, and it can be implemented a few different ways. I chose to use the method outlined by Sheldon Ross in his book Simulation (Fifth Edition), on page 78.
The procedure, and a snippet of the core code used, are both presented in-line below, as an illustration:
| # PROCEDURE, From ROSS: Simulation (5th Edition) Page 78 | |
| # Step 1: Generate Y1, an exponential random variable with rate 1 | |
| Y1 = gen_exponential_distro_rand_variable() | |
| # Step 2: Generate Y2, an exponential random variable with rate 2 | |
| Y2 = gen_exponential_distro_rand_variable() | |
| # Step 3: If Y2 – (Y1 – 1)^2/2 > 0, set Y = Y2 – (Y1 – 1)^2/2, and go to Step 4 (accept) | |
| # Otherwise, go to Step 1 (reject) | |
| subtraction_value = ( math.pow( ( Y1 – 1 ), 2 ) ) / 2 | |
| critical_value = Y2 – subtraction_value | |
| if critical_value > 0: | |
| accept = True | |
| else: | |
| reject = True | |
| # Step 4: Generate a random number on the Uniform Distribution, U, and set: | |
| # Z = Y1 if U <= 1/2 | |
| # Z = Y2 if U >- 1/2if accept == True: | |
| U = random.random() | |
| if (U > 0.5): | |
| Z = Y1 | |
| if (U <= 0.5): | |
| Z = -1.0 * Y1 |
To note: Our Y1 and Y2 values are seeded with variates generated by the exponential distribution, with lambda=1. In much of the literature on the Accept/Reject method, this function is called “g(x)”, and for this example we used the exponential distribution.
It is also important to keep track of how many rejections we get while using the Accept/Reject method. In order to determine the average number of rejections, I ran the algorithm 10 times. The data set it created is shown below:
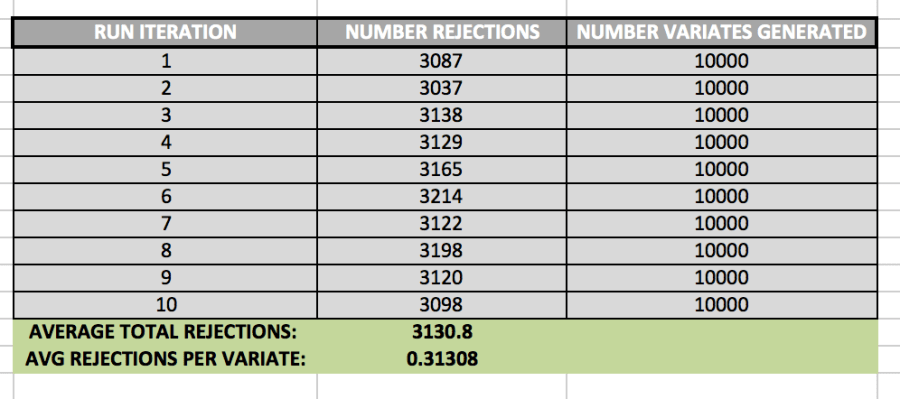
While generating 10,000 variates, the algorithm created 3130.8 rejections, on average. This means that, generally, there was about 1 rejected variate for every 3 valid variates.
Polar-Coordinate Method
In the Polar-Coordinate method, we take advantage of trigonometric properties by generating random variables uniformly distributed over (0, 2pi), and then transforming them into rectangular coordinates. This method, called the Box-Muller Method more formally, is not computationally efficient, however, because it involves use of logs, sines, and cosines—all expensive operations on a CPU.
In order to perform this operation more efficiently, I’ve used the method outlined by Sheldon Ross, on page 83 of his book Simulation (5th Ed.).
Step 1: Generate random numbers, U1 and U2
Step 2: Set V1 = 2U1 – 1, V2 = 2U2 – 1, S = V12 + V22
Step 3: If S > 1, return to Step 1.
Step 4: Return the independent standard normal for two variables, X and Y:
Where:
X = sqrt(-2*log(S)/2) * V1,
Y = sqrt(-2*log(S)/S)*V2
Prior to running the experiment, I expected the Inverse-Transform Method to generate the worst variables themselves, because it only uses an approximation of the Standard Normal CDF, not the CDF itself. I was a little nervous about using an approximation for a CDF with no closed form to generate my inverse function, thinking that while our inverse may deliver results that are more or less reasonable, the resulting data set wouldn’t pass more advanced statistical tests since we are presumably losing precision, through the approximation process. But that said, I also expected its time efficiency to be the best, because we are only calculating logarithm each time we call the inverse function, and this seems to be the only slow operation.
I expected that method 2, Accept/Reject generate the most accurate variables, mostly because of the convincing mathematical proofs describing its validity on pages 77 and 78 of Ross’s Simulation textbook. Intuitively, the proof for this method makes sense, so I expected its data set to look most like something that truly follows the Standard Normal Distribution. From a time efficiency standpoint however, I expected this algorithm to perform 2nd best, because I’m using a logarithm each time I generate an exponential random variable. And with 2 log calls for each run, it seems like this method would be relatively slow, under the assumption that Python’s log function is expensive. (Log calls are used here because we know that –logU is exponential with rate lambda=1. But we need exponential variables generated with rate 1 for each Y variable, Y1 and Y2.)
The Polar Coordinate Method is the most abstract for me, and so I had a hard time seeing exactly why it would generate Standard Normal Random variables, and because of this, I wasn’t sure what to expect of its data set. I took it on faith that it would generate the correct variables, but I didn’t fully understand why. Moreover, I also expected it to perform the worst from a runtime perspective because it involves the most expensive operations: Two Square Roots and Two Log calls for each successful run.
Histograms and Analysis
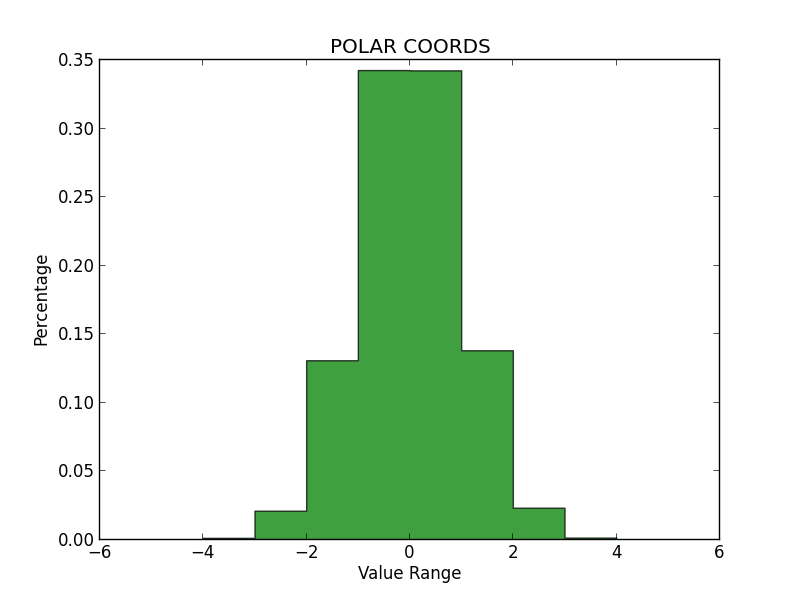
In order to determine whether each method produced qualitatively accurate data, I then plotted histograms for the numbers generated by each. All three histograms can be seen below. On my examination, it appears that the Inverse Transform yielded the least accurate representation, while the Polar Coordinate Method and Accept/Reject were much better, and about equal in validity.
Notably, the Inverse Transform method generated many values beyond the expected range of the Standard Normal (greater than 4.0 and less than -4.0). And while these values are possible, it seems like too many outliers—more than we would see in a true Standard Normal Distribution. Again, I think this is because we are using an approximation for the CDF, rather than the true Inverse of the CDF itself. I had some trouble getting these graphs to sit in-line, so please review the appendix to see them.
Chi-Squared Test
In order to determine wither the numbers generated may be from the Normal Distribution, I ran each method 10 times, and performed a Chi-Square test on each result. The data set can be seen in the tables within the appendix at the back of this document.
From this test, I was able to make the following determinations:
Runtime Analysis
Again, I ran each method 10 times to collect a sample of data with which to analyze runtime.
The average runtimes from 10 tests with n=10,000 were:
This result was indeed surprising. I had expected the Polar Coordinates method to perform the worst, and it did—but only by a very small margin. Moreover, I had expected Inverse Transform to perform the best, and it was only fractions of a microsecond (or nanosecond?) faster than Polar Coordinates on average. I did not expected Accept/Reject to perform so well, but it was by far the fastest overall.
Given these results, I would recommend the Accept/Reject method for anyone who needs to generate Standard Normal Random Variables in Python (at least using my same implementations).
Accept/Reject not only passed the Chi-Square test at the 95% significance level, it also was by far the fastest of the three algorithms. It is roughly comparable to Polar Coordinates on the quality of variables generated, and it beats everything else on speed.
APPENDIX – FIGURES:
Fig 1.1 – Inverse Transform Method Histogram

Fig 1.2 – Polar Coordinates Method HistogramFig 1.3 – Accept/Reject Method Histogram
DATA ANALYSIS TABLES
| INVERSE TRANSFORM | ||||
| RUN ITERATION | CHI SQ | CRIT VALUE, ALPHA=0.05 | NULL HYPOTHESIS | TIME |
| 1 | 29076.10305 | 10233.7489 | REJECT | -7.87E-06 |
| 2 | 28786.13727 | 10233.7489 | REJECT | -5.96E-06 |
| 3 | 29238.94032 | 10233.7489 | REJECT | -6.20E-06 |
| 4 | 27528.91629 | 10233.7489 | REJECT | -8.11E-06 |
| 5 | 28302.76943 | 10233.7489 | REJECT | -5.96E-06 |
| 6 | 28465.05791 | 10233.7489 | REJECT | -5.96E-06 |
| 7 | 28742.14355 | 10233.7489 | REJECT | -6.91E-06 |
| 8 | 29462.56461 | 10233.7489 | REJECT | -5.96E-06 |
| 9 | 28164.87435 | 10233.7489 | REJECT | -6.20E-06 |
| 10 | 28319.68265 | 10233.7489 | REJECT | -6.91E-06 |
| AVG CHI SQ: | 28608.71894 | 10233.7489 | REJECT | |
| AVG TIME SPENT: | -6.60E-06 |
| ACCEPT/REJECT | ||||
| RUN ITERATION | CHI SQ | CRIT VALUE, ALPHA=0.05 | NULL HYPOTHESIS | TIME |
| 1 | 9923.579322 | 10233.7489 | FAIL TO REJECT | -6.91E-06 |
| 2 | 10111.60494 | 10233.7489 | FAIL TO REJECT | -5.01E-06 |
| 3 | 9958.916425 | 10233.7489 | FAIL TO REJECT | -5.01E-06 |
| 4 | 10095.8972 | 10233.7489 | FAIL TO REJECT | -7.15E-06 |
| 5 | 10081.61377 | 10233.7489 | FAIL TO REJECT | -5.96E-06 |
| 6 | 10050.33609 | 10233.7489 | FAIL TO REJECT | -5.01E-06 |
| 7 | 9952.663806 | 10233.7489 | FAIL TO REJECT | -5.01E-06 |
| 8 | 10008.1 | 10233.7489 | FAIL TO REJECT | -5.01E-06 |
| 9 | 9953.795163 | 10233.7489 | FAIL TO REJECT | -6.20E-06 |
| 10 | 10115.71883 | 10233.7489 | FAIL TO REJECT | -5.96E-06 |
| AVG CHI SQ: | 10025.22255 | 10233.7489 | FAIL TO REJECT | |
| AVG TIME SPENT: | -5.72E-06 |
| POLAR COORDINATES | ||||
| RUN ITERATION | CHI SQ | CRIT VALUE, ALPHA=0.05 | NULL HYPOTHESIS | TIME |
| 1 | 9765.748259 | 10233.7489 | FAIL TO REJECT | -5.96E-06 |
| 2 | 9841.898918 | 10233.7489 | FAIL TO REJECT | -4.05E-06 |
| 3 | 10014.11641 | 10233.7489 | FAIL TO REJECT | -5.96E-06 |
| 4 | 10154.0752 | 10233.7489 | FAIL TO REJECT | -7.15E-06 |
| 5 | 10081.61377 | 10233.7489 | FAIL TO REJECT | -7.15E-06 |
| 6 | 9964.385625 | 10233.7489 | FAIL TO REJECT | -5.96E-06 |
| 7 | 9860.196443 | 10233.7489 | FAIL TO REJECT | -4.05E-06 |
| 8 | 9903.479938 | 10233.7489 | FAIL TO REJECT | -1.38E-05 |
| 9 | 10037.27323 | 10233.7489 | FAIL TO REJECT | -7.15E-06 |
| 10 | 10010.40893 | 10233.7489 | FAIL TO REJECT | -5.01E-06 |
| AVG CHI SQ: | 9963.319674 | 10233.7489 | FAIL TO REJECT | |
| AVG TIME SPENT: | -6.63E-06 |
| ACCEPT / REJECT – REJECTIONS | ||
| RUN ITERATION | NUMBER REJECTIONS | NUMBER VARIATES GENERATED |
| 1 | 3087 | 10000 |
| 2 | 3037 | 10000 |
| 3 | 3138 | 10000 |
| 4 | 3129 | 10000 |
| 5 | 3165 | 10000 |
| 6 | 3214 | 10000 |
| 7 | 3122 | 10000 |
| 8 | 3198 | 10000 |
| 9 | 3120 | 10000 |
| 10 | 3098 | 10000 |
| AVERAGE TOTAL REJECTIONS: | 3130.8 | |
| AVG REJECTIONS PER VARIATE: | 0.31308 |
An example of what you’ll find:
| def generate_lcg( num_iterations ): | |
| """ | |
| LCG – generates as many random numbers as requested by user, using a Linear Congruential Generator | |
| LCG uses the formula: X_(i+1) = (aX_i + c) mod m | |
| :param num_iterations: int – the number of random numbers requested | |
| :return: void | |
| """ | |
| # Initialize variables | |
| x_value = 123456789.0 # Our seed, or X_0 = 123456789 | |
| a = 101427 # Our "a" base value | |
| c = 321 # Our "c" base value | |
| m = (2 ** 16) # Our "m" base value | |
| # counter for how many iterations we've run | |
| counter = 0 | |
| # Open a file for output | |
| outFile = open("lgc_output.txt", "wb") | |
| #Perfom number of iterations requested by user | |
| while counter < num_iterations: | |
| # Store value of each iteration | |
| x_value = (a * x_value + c) % m | |
| #Obtain each number in U[0,1) by diving X_i by m | |
| writeValue = str(x_value/m) | |
| # write to output file | |
| outFile.write(writeValue + "\n") | |
| # print "num: " + " " + str(counter) +":: " + str(x_value) | |
| counter = counter+1 | |
| outFile.close() | |
| print("Successfully stored " + str(num_iterations) + " random numbers in file named: 'lgc_output.txt'.") |
I generated some random numbers with a few different generators, some of which I made, and also used the one provided directly by Python. I wanted to see what the best option is for generating random numbers in a few simulations I’m doing.
Grab code here: >> lcg.py <<
It’s commented and can be run by simply invoking Python with: “python lcg.py”
I tried to explain what I was doing at each step to make this clear even for the comparatively un-initiated to the more esoteric statistics at play here, which aren’t totally necessary to know, and really will just be an impediment to __getting_started_now__. I wanted to share something so that people who are practically-minded like me can just jump in, start messing around, and know what to expect. So hopefully with just this code (full repo linked below) and the information presented here, you can start messing around with random number generators if you need to for any reason. (Most common reason would be to seed random variates in a simulation.)
My main goal in posting this is to give anyone with an interest in generating randomness an easy entry into it–with working code for these sort of generators, as it’s somewhat hard to find online, and the details can be a bit opaque, without clear examples of what to expect when you’re testing. So ideally, this will be the total package. Probably not, but hey, giving it a try. If for some reason you need help, feel free to contact me.
Generating truly random numbers is a longstanding problem in math, statistics, and computer science. True randomness requires true entropy, and in many applications—such as generating very large sets of random numbers very quickly—sufficient “true” entropy is difficult or impractical to obtain. (Often, it needs to come from the physical environment, sources such as radioactive decay, etc.) So, instead, we look to algorithmic random number generators for help.
Algorithmically generated random numbers will never be “truly” random precisely because they are generated with a repeatable algorithmic formula. But for purposes such as simulating random events – these “Pseudo-random” numbers can be sufficient. These algorithmic generators take a “seed value” from the environment, or from a user, and use this seed as a variable in their formula to generate as many random-like numbers as a user would like.
Naturally, some of these algorithms are better than others, and hundreds (if not thousands, or more) of them have been designed over the years. The output is always deterministic, and never “truly” random, but the ideal goal is to approximate randomness by generating numbers which:
The best random number generators will pass statistical tests for both uniformity and independence. In this analysis, we will subject three different random number generation algorithms to series of statistical tests and compare the outcomes.
The algorithms we will test are:
The tests each algorithm will be subjected to are:
The exact implementation of each test can be viewed in the linked Python file named: “lcg.py”. The tests can be duplicated by anyone with Python installed on their system by running the command “python lcg.py”.
You can view the file directly on GitHub here: >> lcg.py <<
SUMMARY TABLE (2.1)
|
Algorithm |
X2
TS/Result |
KS TS/Result | Runs
TS/ Result |
Autocorrelation
TS/Result |
| Mersenne Twister (PyRand) |
9.754 Pass all |
0.079213 Pass all |
-1.6605 Reject at 0.8 and 0.9 Pass at 0.95 |
Gap=2: -1.089
Gap=3: -0.924 Gap=5: -0.559 Gap=50: -0.228 All Pass |
| LGC |
6.288 Pass all |
0.0537888 Pass all |
0.5218 Pass all |
Gap=2: 0.1753
Gap=3: -0.394 Gap=5: 0.872 Gap=50: 1.335 Reject at 0.8, Gap=50, others pass |
| LGC, w/ RANDU |
12.336 Pass all |
0.053579 Pass all |
-0.2135 Pass all |
Gap=2: -0.6591
Gap=3: -0.9311 Gap=5: 0.5788 Gap=50:-0.9093 All pass |
The summary table above shows each algorithm tested, and which tests were passed or failed. More detailed output for each test and for each algorithm can be viewed in Tables 1.1 – 1.3 in the appendix to this document.
The Kolmogorov-Smirnov (or KS test) was run at the following levels of significance: .90, 0.95, 0.99. The formulas for the critical value at these significance levels were taken from table of A7 of Discrete-Event System Simulation by Jerry Banks and John S. Carson II. (Formulas for 0.80 could not be found, so I’ve used what was available.)
All other tests were run at the 0.80, 0.90., and 0.95 significance level.
Prior to generating the numbers for each test, I expected Python’s random function to perform the best of all three algorithms tested, mostly because it’s the library random function of one of the world’s most popular programming languages. (Which means: thousands and thousands of code repositories rely on it—many of which are used by commercial and mission critical programs.) But in fact, it performed the worst, failing the Runs Test at both the 0.80 and 0.90 level of significance. These failings are NOT statistically significant at the alpha=0.05 level, but it’s still surprising to see.
I expected the RANDU algorithm to perform the worst, and I thought it would perform especially badly on the autocorrelation test. This is because RANDU is known to have problems, outlined here. Specifically, it is known to produce values which fall along only a specific set of parallel planes (visualization in link above), which means the numbers should NOT be independent, when tested at the right gap lengths. It’s possible that the gap lengths I’ve tested simply missed any of these planes, and as a result—RANDU performed the best of all the algorithms. It’s the only algorithm that didn’t fail any statistical tests at all.
I anticipated the LGC function to perform 2nd best overall, and I was right about that—but the best and worst algorithm were the opposite of what I expected. Mostly, I thought that that Python’s random generator would be nearly perfect, RANDU would be badly flawed, and the LGC would be just okay. Really, the LGC performed admirably: The only test it failed was autocorrelation at the 0.80 confidence level, and that isn’t statistically significant by most measures.
Reviewing the data output into each .txt file directly, I don’t see any discernible patterns in the numbers themselves. And with 10,000 data points, there’s so much output to review that I can see why statistical measures are needed to effectively to determine what’s really going on in the data. It’s probably possible to find a few patterns, specifically related to runs and gap-sequences just by viewing the data directly, but tests are still needed to find out for sure.
With that said, I do think the testing done in this experiment is sufficient, because we have two tests for each measure that matters: 1) Uniformity; 2) Independence. The only improvement I would make for future tests is testing more gap-sequences, and starting them at different points. I’d do this mostly because I know that RANDU should fail gap-sequence tests given the right input, but there would be some trial and error involved in trying to find these sequences naively. So, sometimes, getting into math itself and working with proofs may still be the most effective method. Maybe sometime the old-fashioned way is still best.
TABLE 1.2 – Linear Congruential Generator (X0 = 123456789 )
| Test Name | Sample Size | Confidence Level | Critical Value | Test Statistic Found | Result |
| Chi-Square | 10,000 | 0.80 | 10118.8246 | 6.288 | FAIL TO REJECT null |
| Chi-Square | 10,000 | 0.90 | 10181.6616 | 6.288 | FAIL TO REJECT null |
| Chi-Square | 10,000 | 0.95 | 10233.7489 | 6.288 | FAIL TO REJECT null |
| Kolmogorov-Smirnov | 100 | 0.90 | 0.122 | 0.05378 | FAIL TO REJECT null |
| Kolmogorov-Smirnov | 100 | 0.95 | 0.136 | 0.05378 | FAIL TO REJECT null |
| Kolmogorov-Smirnov | 100 | 0.99 | 0.163 | 0.05378 | FAIL TO REJECT null |
| Runs Test | 10,000 | 0.80 | 1.282 | 0.521889 | FAIL TO REJECT null |
| Runs Test | 10,000 | 0.90 | 1.645 | 0.521889 | FAIL TO REJECT null |
| Runs Test | 10,000 | 0.95 | 1.96 | 0.521889 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=2 |
10,000 | 0.80 | 1.282 | 0.175377 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=2 |
10,000 | 0.90 | 1.645 | 0.1753777 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=2 |
10,000 | 0.95 | 1.96 | 0.1753777 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=3 |
10,000 | 0.8 | 1.282 | -0.39487 | FAIL TO REJECT null
|
| Autocorrelation,
GapSize=3 |
10,000 | 0.9 | 1.645 | -0.39487 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=3 |
10,000 | 0.95 | 1.96 | -0.39487 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=5 |
10,000 | 0.8 | 1.282 | 0.872668 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=5 |
10,000 | 0.9 | 1.645 | 0.872668 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=5 |
10,000 | 0.95 | 1.96 | 0.872668 | FAIL TO REJECT
null |
| Autocorrelation,
GapSize=50 |
10,000 | 0.8 | 1.282 | 1.3352 | REJECT null |
| Autocorrelation,
GapSize=50 |
10,000 | 0.9 | 1.645 | 1.3352 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=50 |
10,000 | 0.95 | 1.96 | 1.3352 | FAIL TO REJECT null |
| Test Name | Sample Size | Confidence Level | Critical Value | Test Statistic Found | Result |
END TABLE 1.2
TABLE 1.3 – Linear Congruential Generator with RANDU initial settings
| Test Name | Sample Size | Confidence Level | Critical Value | Test Statistic Found | Result |
| Chi-Square | 10,000 | 0.80 | 10118.8246 | 12.336 | FAIL TO REJECT null |
| Chi-Square | 10,000 | 0.90 | 10181.6616 | 12.336 | FAIL TO REJECT null |
| Chi-Square | 10,000 | 0.95 | 10233.7489 | 12.336 | FAIL TO REJECT null |
| Kolmogorov-Smirnov | 100 | 0.90 | 0.122 | 0.053579 | FAIL TO REJECT null |
| Kolmogorov-Smirnov | 100 | 0.95 | 0.136 | 0.053579 | FAIL TO REJECT null |
| Kolmogorov-Smirnov | 100 | 0.99 | 0.163 | 0.053579 | FAIL TO REJECT null |
| Runs Test | 10,000 | 0.8 | 1.282 | -0.21350 | FAIL TO REJECT null |
| Runs Test | 10,000 | 0.9 | 1.645 | -0.21350 | FAIL TO REJECT null |
| Runs Test | 10,000 | 0.95 | 1.96 | -0.21350 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=2 |
10,000 | 0.8 | 1.282 | -0.65918 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=2 |
10,000 | 0.9 | 1.645 | -0.65918 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=2 |
10,000 | 0.95 | 1.96 | -0.65918 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=3 |
10,000 | 0.8 | 1.282 | -0.93113 | FAIL TO REJECT null
|
| Autocorrelation,
GapSize=3 |
10,000 | 0.9 | 1.645 | -0.93113 | FAIL TO REJECT null
|
| Autocorrelation,
GapSize=3 |
10,000 | 0.95 | 1.96 | -0.93113 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=5 |
10,000 | 0.80 | 1.282 | 0.378881 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=5 |
10,000 | 0.90 | 1.645 | 0.378881 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=5 |
10,000 | 0.95 | 1.96 | 0.378881 | FAIL TO REJECT
null |
| Autocorrelation,
GapSize=50 |
10,000 | 0.80 | 1.282 | -0.90937 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=50 |
10,000 | 0.90 | 1.645 | -0.90937 | FAIL TO REJECT null |
| Autocorrelation,
GapSize=50 |
10,000 | 0.95 | 1.96 | -0.90937 | FAIL TO REJECT null |
| Test Name | Sample Size | Confidence Level | Critical Value | Test Statistic Found | Result |
END TABLE 1.3
Table 2.1 – Test Result Comparisons
|
Algorithm |
X2
TS/Result |
KS TS/Result | Runs
TS/ Result |
Autocorrelation
TS/Result |
| Mersenne Twister (PyRand) |
9.754 Pass all |
0.079213 Pass all |
-1.6605 Reject at 0.8 and 0.9 Pass at 0.95 |
Gap=2: -1.089
Gap=3: -0.924 Gap=5: -0.559 Gap=50: -0.228 All Pass |
| LGC |
6.288 Pass all |
0.0537888 Pass all |
0.5218 Pass all |
Gap=2: 0.1753
Gap=3: -0.394 Gap=5: 0.872 Gap=50: 1.335 Reject at 0.8, Gap=50, others pass |
| LGC, w/ RANDU |
12.336 Pass all |
0.053579 Pass all |
-0.2135 Pass all |
Gap=2: -0.6591
Gap=3: -0.9311 Gap=5: 0.5788 Gap=50:-0.9093 All pass |
You must be logged in to post a comment.